Abstract
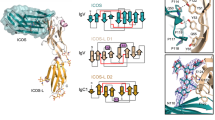
The dimeric cell-surface glycoprotein CD8 is crucial to the positive selection of cytotoxic T cells in the thymus1. The homodimer CD8αα or the heterodimer αβ stabilizes the interaction of the T-cell antigen receptor (TCR) with major histocompatibility complex (MHC) class I/peptide by binding to the class I molecule2. Here we report the crystal structure at 2.7Å resolution of a complex between CD8αα and the human MHC molecule HLA-A2, which is associated with peptide. CD8αα binds one HLA-A2/peptide molecule, interfacing with the α2 and α3 domains of HLA-A2 and also contacting β2-microglobulin. A flexible loop of the α3 domain (residues 223–229) is clamped between the complementarity-determining region (CDR)-like loops of the two CD8 subunits in the classic manner of an antibody–antigen interaction, precluding the binding of a second MHC molecule. The position of the α3 domain is different from that in uncomplexed HLA-A2 (refs 3, 4), being most similar to that in the TCR/Tax/HLA-A2 complex5, but no conformational change extends to the MHC/peptide surface presented for TCR recognition. Although these shifts in α3 may provide a synergistic modulation of affinity, the binding of CD8 to MHC is clearly consistent with an avidity-based contribution from CD8 to TCR–peptide–MHC interactions.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Zamoyska, R. The CD8 coreceptor revisited: one chain good, two chains better. Immunity 1, 243–246 (1994).
Norment, A. M., Salter, R. D., Parham, P., Engelhard, V. H. & Littman, D. R. Cell–cell adhesion mediated by CD8 and MHC class I molecules. Nature 336, 79–81 (1988).
Madden, D. R., Garboczi, D. N. & Wiley, D. C. The antigenic identity of peptide–MHC complexes: a comparison of the conformations of five viral peptides presented by HLA-A2. Cell 75, 693–708 (1993).
Bjorkman, P. J. et al. Structure of the human class I histocompatibility antigen, HLA-A2. Nature 329, 506–512 (1987).
Garboczi, D. N. et al. Structure of the complex between human T-cell receptor, viral peptide and HLA-A2. Nature 384, 134–141 (1996).
Leahy, D. J., Axel, R. & Hendrickson, W. A. Crystal structure of a soluble form of the human T cell coreceptor CD8 at 2.6Å resolution. Cell 68, 1145–1162 (1992).
Madden, D. R. The three-dimensional structure of peptide-MHC complexes. Annu. Rev. Immunol. 13, 587–622 (1995).
Kirszbaum, L., Sharpe, J. A., Goss, N., Lahnstein, J. & Walker, I. D. The alpha-chain of murine CD8 lacks an invariant Ig-like disulfide bond but contains a unique intrachain loop instead. J. Immunol. 142, 3931–3936 (1989).
Salter, R. D. et al. Polymorphism in the α3 domain of HLA-A molecules affects binding to CD8. Nature 338, 345–347 (1989).
Salter, R. D. et al. Abinding site for the T-cell co-receptor CD8 on the α3 domain of HLA-A2. Nature 345, 41–46 (1990).
Sun, J., Leahy, D. J. & Kavathas, P. B. Interaction between CD8 and major histocompatibility complex (MHC) class I mediated by multiple contact surface that include the alpha 2 and alpha 3 domains of MHC class I. J. Exp. Med. 182, 1275–1280 (1995).
Wilson, I. A. & Stanfield, R. L. Antibody-antigen interaction: new structures and new conformational changes. Curr. Opin. Struct. Biol. 4, 857–867 (1994).
Clackson, T. & Wells, J. A. Ahot spot of binding energy in a hormone-receptor interface. Science 267, 383–386 (1995).
Garrett, T. P., Saper, M. A., Bjorkman, P. J., Strominger, J. L. & Wiley, D. C. Specificity pockets for the side chains of peptide antigens in HLA-Aw68. Nature 342, 692–696 (1989).
Giblin, P. A., Leahy, D. J., Mennone, J. & Kavathas, P. B. The role of charge and multiple faces of the CD8 alpha/alpha homodimer in binding to major histocompatibility complex class I molecules: support for a bivalent model. Proc. Natl Acad. Sci. USA 91, 1716–1720 (1994).
Luescher, I. F. et al. CD8 modulation of T-cell antigen receptor–ligand interactions on living cytotoxic T lymphocytes. Nature 373, 353–356 (1995).
Wheeler, C. J., von Hoegen, P. & Parnes, J. R. An immunological role for the CD8 β-chain. Nature 357, 247–249 (1992).
Garcia, K. C. et al. CD8 enhances formation of stable T-cell receptor./MHC class I molecule complexes. Nature 384, 577–581 (1996).
Reid, S. W. et al. Production and crystallization of MHC class I B allele single peptide complexes. FEBS Lett. 383, 119–123 (1996).
Otwinowski, Z. & Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997).
Navaza, J. AMoRe: an automated package for molecular replacement. Acta Crystallogr. A 50, 157–163 (1994).
Brünger, A. T. XPLOR Version 3.1: A System for X-ray Crystallography and NMR (Yale Univ. Press, New Haven, CT, (1992)).
Jones, T. A., Zou, J. Y., Cowan, S. W. & Kjeldgaard, M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47, 110–119 (1991).
Kabsch, W. & Sander, C. Dictionary of protein secondary structure: Pattern recognition of hydrogen-bonded and geometrical features. Biopolymers 22, 2577–2637 (1983).
Kraulis, P. J. MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr. 24, 946–950 (1991).
Merritt, E. A. & Murphy, M. E. P. Raster3D version 2.0. A program for photorealistic molecular graphics. Acta Crystallogr. D 50, 869–873 (1994).
Smith, K. J. et al. Bound water structure and polymorphic amino acids act together to allow the binding of different peptides to MHC class I HLA-B53. Immunity 4, 215–228 (1996).
Stuart, D. I., Levine, M., Muirhead, H. & Stammers, D. K. The crystal structure of cat pyruvate kinase at a resolution of 2.6Å. J. Mol. Biol. 134, 109–142 (1979).
Acknowledgements
We are grateful to the late Alan Williams for discussions that inspired this work. We thank R. Bryan, K. Measures and R. Esnouf for computing facilities and programs; K. Harlos for assistance with X-ray data collection, S. Lee for help in the preparation of figures; Z. Rao for assistance in crystallization trials; D. Wiley and D. Garboczi for pre-release coordinates of the TCR/Tax/HLA-A2 complex; and C. O'Callahan, V. Cerundolo, D. Garboczi, G. Harcourt and B. Willcox for help and advice. This work was funded by the MRC. The Oxford Centre for Molecular Sciences is supported by the BBSRC, EPSRC and MRC. J.T. was supported by an EMBO fellowship, E.Y.J. by the Royal Society, and D.I.S. and A.J.M. by the MRC.
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Gao, G., Tormo, J., Gerth, U. et al. Crystal structure of the complex between human CD8αα and HLA-A2. Nature 387, 630–634 (1997). https://doi.org/10.1038/42523
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/42523
This article is cited by
-
Evolution and molecular interactions of major histocompatibility complex (MHC)-G, -E and -F genes
Cellular and Molecular Life Sciences (2022)
-
Modulation of innate and adaptive immunity by cytomegaloviruses
Nature Reviews Immunology (2020)
-
Structural understanding of T cell receptor triggering
Cellular & Molecular Immunology (2020)
-
Challenging immunodominance of influenza-specific CD8+ T cell responses restricted by the risk-associated HLA-A*68:01 allomorph
Nature Communications (2019)
-
Defining the structural basis for human alloantibody binding to human leukocyte antigen allele HLA-A*11:01
Nature Communications (2019)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.