Abstract
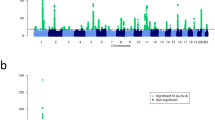
To gain further insight into the genetic architecture of psoriasis, we conducted a meta-analysis of 3 genome-wide association studies (GWAS) and 2 independent data sets genotyped on the Immunochip, including 10,588 cases and 22,806 controls. We identified 15 new susceptibility loci, increasing to 36 the number associated with psoriasis in European individuals. We also identified, using conditional analyses, five independent signals within previously known loci. The newly identified loci shared with other autoimmune diseases include candidate genes with roles in regulating T-cell function (such as RUNX3, TAGAP and STAT3). Notably, they included candidate genes whose products are involved in innate host defense, including interferon-mediated antiviral responses (DDX58), macrophage activation (ZC3H12C) and nuclear factor (NF)-κB signaling (CARD14 and CARM1). These results portend a better understanding of shared and distinctive genetic determinants of immune-mediated inflammatory disorders and emphasize the importance of the skin in innate and acquired host defense.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout
Similar content being viewed by others
References
Nestle, F.O., Kaplan, D.H. & Barker, J. Psoriasis. N. Engl. J. Med. 361, 496–509 (2009).
Elder, J.T. et al. Molecular dissection of psoriasis: integrating genetics and biology. J. Invest. Dermatol. 130, 1213–1226 (2010).
Ellinghaus, E. et al. Genome-wide association study identifies a psoriasis susceptibility locus at TRAF3IP2. Nat. Genet. 42, 991–995 (2010).
Nair, R.P. et al. Genome-wide scan reveals association of psoriasis with IL-23 and NF-κB pathways. Nat. Genet. 41, 199–204 (2009).
Strange, A. et al. A genome-wide association study identifies new psoriasis susceptibility loci and an interaction between HLA-C and ERAP1. Nat. Genet. 42, 985–990 (2010).
Stuart, P.E. et al. Genome-wide association analysis identifies three psoriasis susceptibility loci. Nat. Genet. 42, 1000–1004 (2010).
Sun, L.D. et al. Association analyses identify six new psoriasis susceptibility loci in the Chinese population. Nat. Genet. 42, 1005–1009 (2010).
Zhang, X.J. et al. Psoriasis genome-wide association study identifies susceptibility variants within LCE gene cluster at 1q21. Nat. Genet. 41, 205–210 (2009).
de Cid, R. et al. Deletion of the late cornified envelope LCE3B and LCE3C genes as a susceptibility factor for psoriasis. Nat. Genet. 41, 211–215 (2009).
Ellinghaus, D. et al. Combined analysis of genome-wide association studies for Crohn disease and psoriasis identifies seven shared susceptibility loci. Am. J. Hum. Genet. 90, 636–647 (2012).
Cotsapas, C. et al. Pervasive sharing of genetic effects in autoimmune disease. PLoS Genet. 7, e1002254 (2011).
Cortes, A. & Brown, M.A. Promise and pitfalls of the Immunochip. Arthritis Res. Ther. 13, 101 (2011).
Trynka, G. et al. Dense genotyping identifies and localizes multiple common and rare variant association signals in celiac disease. Nat. Genet. 43, 1193–1201 (2011).
Feng, B.J. et al. Multiple loci within the major histocompatibility complex confer risk of psoriasis. PLoS Genet. 5, e1000606 (2009).
Zheng, H.F. et al. Variants in MHC, LCE and IL12B have epistatic effects on psoriasis risk in Chinese population. J. Dermatol. Sci. 61, 124–128 (2011).
Riveira-Munoz, E. et al. Meta-analysis confirms the LCE3C_LCE3B deletion as a risk factor for psoriasis in several ethnic groups and finds interaction with HLA-Cw6. J. Invest. Dermatol. 131, 1105–1109 (2011).
Jordan, C.T. et al. PSORS2 is due to mutations in CARD14. Am. J. Hum. Genet. 90, 784–795 (2012).
Jordan, C.T. et al. Rare and common variants in CARD14, encoding an epidermal regulator of NF-κB, in psoriasis. Am. J. Hum. Genet. 90, 796–808 (2012).
Di Meglio, P. et al. The IL23R R381Q gene variant protects against immune-mediated diseases by impairing IL-23–induced Th17 effector response in humans. PLoS ONE 6, e17160 (2011).
Cal, S. et al. Identification and characterization of human polyserase-3, a novel protein with tandem serine-protease domains in the same polypeptide chain. BMC Biochem. 7, 9 (2006).
Dubois, P.C. et al. Multiple common variants for celiac disease influencing immune gene expression. Nat. Genet. 42, 295–302 (2010).
Zhernakova, A. et al. Meta-analysis of genome-wide association studies in celiac disease and rheumatoid arthritis identifies fourteen non-HLA shared loci. PLoS Genet. 7, e1002004 (2011).
Franke, A. et al. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn's disease susceptibility loci. Nat. Genet. 42, 1118–1125 (2010).
Gudjonsson, J.E. et al. Assessment of the psoriatic transcriptome in a large sample: additional regulated genes and comparisons with in vitro models. J. Invest. Dermatol. 130, 1829–1840 (2010).
Ding, J. et al. Gene expression in skin and lymphoblastoid cells: refined statistical method reveals extensive overlap in cis-eQTL signals. Am. J. Hum. Genet. 87, 779–789 (2010).
Mason, C.C. et al. Bimodal distribution of RNA expression levels in human skeletal muscle tissue. BMC Genomics 12, 98 (2011).
Song, M.Y., Kim, H.E., Kim, S., Choi, I.H. & Lee, J.K. SNP-based large-scale identification of allele-specific gene expression in human B cells. Gene 493, 211–218 (2012).
Andrés, A.M. et al. Balancing selection maintains a form of ERAP2 that undergoes nonsense-mediated decay and affects antigen presentation. PLoS Genet. 6, e1001157 (2010).
Grjibovski, A.M., Olsen, A.O., Magnus, P. & Harris, J.R. Psoriasis in Norwegian twins: contribution of genetic and environmental effects. J. Eur. Acad. Dermatol. Venereol. 21, 1337–1343 (2007).
Najarian, D.J. & Gottlieb, A.B. Connections between psoriasis and Crohn's disease. J. Am. Acad. Dermatol. 48, 805–821 quiz 822–824 (2003).
Ludvigsson, J.F., Lindelof, B., Zingone, F. & Ciacci, C. Psoriasis in a nationwide cohort study of patients with celiac disease. J. Invest. Dermatol. 131, 2010–2016 (2011).
Modlin, R.L. Innate immunity: ignored for decades, but not forgotten. J. Invest. Dermatol. 132, 882–886 (2012).
Nakatsuji, T. & Gallo, R.L. Antimicrobial peptides: old molecules with new ideas. J. Invest. Dermatol. 132, 887–895 (2012).
Wölfle, U., Martin, S., Emde, M. & Schempp, C. Dermatology in the Darwin anniversary. Part 2: Evolution of the skin-associated immune system. J. Dtsch. Dermatol. Ges. 7, 862–869 (2009).
Capon, F., Burden, A.D., Trembath, R.C. & Barker, J.N. Psoriasis and other complex trait dermatoses: from loci to functional pathways. J. Invest. Dermatol. 132, 915–922 (2012).
Gudjonsson, J.E. & Elder, J.T. Psoriasis. in Dermatology in General Medicine Vol. 1 (eds. Goldsmith, L. et al.) 197–231 (McGraw-Hill, New York, 2012).
Garber, K. Psoriasis: from bed to bench and back. Nat. Biotechnol. 29, 563–566 (2011).
Vilhais-Neto, G.C. et al. Rere controls retinoic acid signalling and somite bilateral symmetry. Nature 463, 953–957 (2010).
Ferby, I. et al. Mig6 is a negative regulator of EGF receptor–mediated skin morphogenesis and tumor formation. Nat. Med. 12, 568–573 (2006).
Amler, L.C. et al. Identification and characterization of novel genes located at the t(1;15)(p36.2;q24) translocation breakpoint in the neuroblastoma cell line NGP. Genomics 64, 195–202 (2000).
Djuretic, I.M. et al. Transcription factors T-bet and Runx3 cooperate to activate Ifng and silence Il4 in T helper type 1 cells. Nat. Immunol. 8, 145–153 (2007).
Shiraishi, N. et al. Identification and characterization of three novel β1,3-N-acetylglucosaminyltransferases structurally related to the β1,3-galactosyltransferase family. J. Biol. Chem. 276, 3498–3507 (2001).
Togayachi, A. β3GnT2 (B3GNT2), a major polylactosamine synthase: analysis of B3GNT2-deficient mice. Methods Enzymol. 479, 185–204 (2010).
Grindstaff, K.K. et al. Sec6/8 complex is recruited to cell-cell contacts and specifies transport vesicle delivery to the basal-lateral membrane in epithelial cells. Cell 93, 731–740 (1998).
Biswas, P.S. et al. Phosphorylation of IRF4 by ROCK2 regulates IL-17 and IL-21 production and the development of autoimmunity in mice. J. Clin. Invest. 120, 3280–3295 (2010).
Mudter, J. et al. IRF4 regulates IL-17A promoter activity and controls RORγt-dependent Th17 colitis in vivo. Inflamm. Bowel Dis. 17, 1343–1358 (2011).
Huber, M. et al. IRF4 is essential for IL-21–mediated induction, amplification, and stabilization of the Th17 phenotype. Proc. Natl. Acad. Sci. USA 105, 20846–20851 (2008).
Bowcock, A.M. et al. Insights into psoriasis and other inflammatory diseases from large-scale gene expression studies. Hum. Mol. Genet. 10, 1793–1805 (2001).
Chang, I.F. & Hsiao, H.Y. Induction of RhoGAP and pathological changes characteristic of Alzheimer's disease by UAHFEMF discharge in rat brain. Curr. Alzheimer Res. 2, 559–569 (2005).
Gotoh, K. et al. Selective control of type I IFN induction by the Rac activator DOCK2 during TLR-mediated plasmacytoid dendritic cell activation. J. Exp. Med. 207, 721–730 (2010).
Ippagunta, S.K. et al. The inflammasome adaptor ASC regulates the function of adaptive immune cells by controlling Dock2-mediated Rac activation and actin polymerization. Nat. Immunol. 12, 1010–1016 (2011).
Loo, Y.M. & Gale, M. Immune signaling by RIG-I–like receptors. Immunity 34, 680–692 (2011).
Cui, X.F., Imaizumi, T., Yoshida, H., Borden, E.C. & Satoh, K. Retinoic acid–inducible gene-I is induced by interferon-γ and regulates the expression of interferon-γ stimulated gene 15 in MCF-7 cells. Biochem. Cell Biol. 82, 401–405 (2004).
Negishi, H. et al. A critical link between Toll-like receptor 3 and type II interferon signaling pathways in antiviral innate immunity. Proc. Natl. Acad. Sci. USA 105, 20446–20451 (2008).
Patel, S., Xi, Z.F., Seo, E.Y., McGaughey, D. & Segre, J.A. Klf4 and corticosteroids activate an overlapping set of transcriptional targets to accelerate in utero epidermal barrier acquisition. Proc. Natl. Acad. Sci. USA 103, 18668–18673 (2006).
Feinberg, M.W. et al. Kruppel-like factor 4 is a mediator of proinflammatory signaling in macrophages. J. Biol. Chem. 280, 38247–38258 (2005).
Liang, J. et al. A novel CCCH-zinc finger protein family regulates proinflammatory activation of macrophages. J. Biol. Chem. 283, 6337–6346 (2008).
Nagarajan, P. et al. Ets1 blocks terminal differentiation of keratinocytes and induces expression of matrix metalloproteases and innate immune mediators. J. Cell Sci. 123, 3566–3575 (2010).
Zamisch, M. et al. The transcription factor Ets1 is important for CD4 repression and Runx3 up-regulation during CD8 T cell differentiation in the thymus. J. Exp. Med. 206, 2685–2699 (2009).
Moisan, J., Grenningloh, R., Bettelli, E., Oukka, M. & Ho, I.C. Ets-1 is a negative regulator of Th17 differentiation. J. Exp. Med. 204, 2825–2835 (2007).
Sakamoto, H. et al. A Janus kinase inhibitor, JAB, is an interferon-γ–inducible gene and confers resistance to interferons. Blood 92, 1668–1676 (1998).
Tanaka, K. et al. Loss of suppressor of cytokine signaling 1 in helper T cells leads to defective Th17 differentiation by enhancing antagonistic effects of IFN-γ on STAT3 and Smads. J. Immunol. 180, 3746–3756 (2008).
Piganis, R.A. et al. Suppressor of cytokine signaling (SOCS) 1 inhibits type I interferon (IFN) signaling via the interferon α receptor (IFNAR1)-associated tyrosine kinase Tyk2. J. Biol. Chem. 286, 33811–33818 (2011).
Sano, S. et al. Stat3 links activated keratinocytes and immunocytes required for development of psoriasis in a novel transgenic mouse model. Nat. Med. 11, 43–49 (2005).
Harris, T.J. et al. Cutting edge: an in vivo requirement for STAT3 signaling in TH17 development and TH17-dependent autoimmunity. J. Immunol. 179, 4313–4317 (2007).
Wei, L., Laurence, A. & O'Shea, J.J. New insights into the roles of Stat5a/b and Stat3 in T cell development and differentiation. Semin. Cell Dev. Biol. 19, 394–400 (2008).
Blonska, M. & Lin, X. NF-κB signaling pathways regulated by CARMA family of scaffold proteins. Cell Res. 21, 55–70 (2011).
Kersh, E.N. Impaired memory CD8 T cell development in the absence of methyl-CpG–binding domain protein 2. J. Immunol. 177, 3821–3826 (2006).
Faili, A. et al. Induction of somatic hypermutation in immunoglobulin genes is dependent on DNA polymerase iota. Nature 419, 944–947 (2002).
Inouye, M. et al. An immune response network associated with blood lipid levels. PLoS Genet. 6, pii e1001113 (2010).
Wichmann, H.E., Gieger, C. & Illig, T. KORA-gen—resource for population genetics, controls and a broad spectrum of disease phenotypes. Gesundheitswesen 67 suppl 1, S26–S30 (2005).
1000 Genomes Project Consortium. A map of human genome variation from population-scale sequencing. Nature 467, 1061–1073 (2010).
Giannoulatou, E., Yau, C., Colella, S., Ragoussis, J. & Holmes, C.C. GenoSNP: a variational Bayes within-sample SNP genotyping algorithm that does not require a reference population. Bioinformatics 24, 2209–2214 (2008).
Howie, B., Fuchsberger, C., Stephens, M., Marchini, J. & Abecasis, G. Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat. Genet. 44, 955–959 (2012).
Marchini, J., Howie, B., Myers, S., McVean, G. & Donnelly, P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat. Genet. 39, 906–913 (2007).
Howie, B.N., Donnelly, P. & Marchini, J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 5, e1000529 (2009).
Altshuler, D.M. et al. Integrating common and rare genetic variation in diverse human populations. Nature 467, 52–58 (2010).
Purcell, S. et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575 (2007).
Kang, H.M. et al. Variance component model to account for sample structure in genome-wide association studies. Nat. Genet. 42, 348–354 (2010).
Devlin, B. & Roeder, K. Genomic control for association studies. Biometrics 55, 997–1004 (1999).
Freedman, M.L. et al. Assessing the impact of population stratification on genetic association studies. Nat. Genet. 36, 388–393 (2004).
Acknowledgements
Major support for this study was provided by the US National Institutes of Health, the Wellcome Trust and the German Research Foundation. We thank J.C. Barrett for contribution to the design of the Immunochip and helpful analytical discussion, as well as E. Gray, S. Bumpstead, D. Simpkin and the staff of the Wellcome Trust Sanger Institute Sample Management and Genotyping teams for their genotyping and analytical contributions. We acknowledge use of the British 1958 Birth Cohort DNA collection, funded by the UK Medical Research Council (G0000934) and the Wellcome Trust (068545/Z/02), and the UK National Blood Service controls, funded by the Wellcome Trust. We acknowledge CASP for the contribution of GWAS data, as well as the provision of control DNA samples by the Cooperative Research in the Region of Augsburg (KORA) and Heinz-Nixdorf Recall (Risk Factors, Evaluation of Coronary Calcification and Lifestyle) study (HNR) and genotyping data generated by the Dietary, Lifestyle and Genetic determinants of Obesity and Metabolic syndrome (DILGOM) Consortium. We thank the Barbara and Neal Henschel Charitable Foundation for their support of the National Psoriasis Victor Henschel BioBank. We acknowledge the Genetic repository in Ireland for Psoriasis and Psoriatic Arthritis (GRIPPsA), the Irish blood transfusion service/Trinity College Dublin Biobank and the Dublin Centre for Clinical Research (funded by the Health Research Board and the Wellcome Trust). A detailed list of contributing consortia and relevant funding support is provided in the Supplementary Note.
Author information
Authors and Affiliations
Consortia
Contributions
J.T.E., R.C.T. and G.R.A. designed and directed the study. R.P.N., M.W., J.D., J.J.V., J.T.E., F.C., J.N.W.N.B., M.H.A., C.H.S., A.D.B., C.E.M.G., A.R., J. Kere, X.E., W.W., J. Worthington, R.T.-A., M.S., G.N., L.S., R.M., M.J.C., J.S., A.F., S.W., S.K., K.K., T.E., A.M., A.M.B., G.G.K., D.D.G., P.R., U.M., F.O.N., A.H., J. Winkelmann, S.S., C.W., C.L., S.E., R.A., V.C., C.F.R., H.B., H.W.L. and H.E.W. contributed to sample collection and phenotyping. J. Knight coordinated the samples and data sets for the Genetic Analysis of Psoriasis (GAP) consortium. J.T.E. coordinated the PAGE samples and data sets. P.D., A.S., G.B., R.D.P., D.V. and C.C.A.S. contributed to the design of the Immunochip. J. Knight, P.E.S., G.R.A. and H.M.K. advised on the statistical analysis. C.L., S.E., R.A., H.B., E.E., P.H. and R.P.N. performed genotyping. E.E., S.L.S., L.C.T. and H.M.K. performed the genotype calling. S.L.S., L.C.T., Y.L. and J.D. performed genotype imputation and statistical analysis. F.C., J.N.W.N.B., J.E.G., T.T., J.T.E. and A.F. prepared Box 1. L.C.T., S.L.S., F.C. and J.T.E. drafted the manuscript and prepared the figures and tables. E.E., J.E.G., J. Knight, P.E.S., R.P.N., R.C.T., T.T., G.R.A., J.N.W.N.B. and A.F. edited and revised the manuscript. All authors approved the final draft.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Tables 1–10, Supplementary Figures 1–5 and Supplementary Note (PDF 2509 kb)
Rights and permissions
About this article
Cite this article
Tsoi, L., Spain, S., Knight, J. et al. Identification of 15 new psoriasis susceptibility loci highlights the role of innate immunity. Nat Genet 44, 1341–1348 (2012). https://doi.org/10.1038/ng.2467
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.2467
This article is cited by
-
RBP–RNA interactions in the control of autoimmunity and autoinflammation
Cell Research (2023)
-
Immunosuppression causes dynamic changes in expression QTLs in psoriatic skin
Nature Communications (2023)
-
Disentangling the complexity of psoriasis in the post-genome-wide association era
Genes & Immunity (2023)
-
Could NCOA5 a novel candidate gene for multiple sclerosis susceptibility?
Molecular Biology Reports (2023)
-
A common human MLKL polymorphism confers resistance to negative regulation by phosphorylation
Nature Communications (2023)