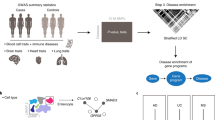
Abstract
Rheumatic diseases have complex aetiologies that are not fully understood, which makes the study of pathogenic mechanisms in these diseases a challenge for researchers. Next-generation sequencing (NGS) and related omics technologies, such as transcriptomics, epigenomics and genomics, provide an unprecedented genome-wide view of gene expression, environmentally responsive epigenetic changes and genetic variation. The integrated application of NGS technologies to samples from carefully phenotyped clinical cohorts of patients has the potential to solve remaining mysteries in the pathogenesis of several rheumatic diseases, to identify new therapeutic targets and to underpin a precision medicine approach to the diagnosis and treatment of rheumatic diseases. This Review provides an overview of the NGS technologies available, showcases important advances in rheumatic disease research already powered by these technologies and highlights NGS approaches that hold particular promise for generating new insights and advancing the field.
Key points
-
Next-generation sequencing (NGS) technologies have the potential to provide insight into the interaction between environmental factors and genetics in the pathogenesis of rheumatic diseases.
-
Transcriptomic studies have revealed disease-related pathways and novel pathogenic cell types in rheumatic diseases.
-
Epigenomic studies have revealed memory-related phenomena that might help to explain the chronicity of disease and have linked enhancers harbouring disease-associated allelic variants with target genes.
-
Whole-genome sequencing and exome sequencing have revealed causal mutations in rare Mendelian autoinflammatory diseases.
-
NGS approaches will substantially contribute to the application of precision medicine in rheumatology.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Banchereau, R., Cepika, A. M., Banchereau, J. & Pascual, V. Understanding human autoimmunity and autoinflammation through transcriptomics. Annu. Rev. Immunol. 35, 337–370 (2017).
Ermann, J., Rao, D. A., Teslovich, N. C., Brenner, M. B. & Raychaudhuri, S. Immune cell profiling to guide therapeutic decisions in rheumatic diseases. Nat. Rev. Rheumatol. 11, 541–551 (2015).
Byron, S. A., Van Keuren-Jensen, K. R., Engelthaler, D. M., Carpten, J. D. & Craig, D. W. Translating RNA sequencing into clinical diagnostics: opportunities and challenges. Nat. Rev. Genet. 17, 257–271 (2016).
Davis, M. M. & Brodin, P. Rebooting human immunology. Annu. Rev. Immunol. 36, 843–864 (2018).
Donlin, L. T. et al. Methods for high-dimensional analysis of cells dissociated from cryopreserved synovial tissue. Arthritis Res. Ther. 20, 139 (2018).
Der, E. et al. Tubular cell and keratinocyte single-cell transcriptomics applied to lupus nephritis reveal type I IFN and fibrosis relevant pathways. Nat. Immunol. (in the press).
Rao, D. A. et al. A protocol for single-cell transcriptomics from cryopreserved renal tissue and urine for the Accelerating Medicine Partnership (AMP) RA/SLE network. Preprint at bioRxiv https://www.biorxiv.org/content/10.1101/275859v1 (2018).
Eikrem, O. et al. Transcriptome sequencing (RNAseq) enables utilization of formalin-fixed, paraffin-embedded biopsies with clear cell renal cell carcinoma for exploration of disease biology and biomarker development. PLOS ONE 11, e0149743 (2016).
Banchereau, R. et al. Personalized immunomonitoring uncovers molecular networks that stratify lupus patients. Cell 165, 1548–1550 (2016).
Dennis, G. Jr. et al. Synovial phenotypes in rheumatoid arthritis correlate with response to biologic therapeutics. Arthritis Res. Ther. 16, R90 (2014).
Costa-Silva, J., Domingues, D. & Lopes, F. M. RNA-Seq differential expression analysis: An extended review and a software tool. PLOS ONE 12, e0190152 (2017).
Wang, X. et al. Three-dimensional intact-tissue sequencing of single-cell transcriptional states. Science 361, eaat5691 (2018).
Carlucci, P. M. et al. Neutrophil subsets and their gene signature associate with vascular inflammation and coronary atherosclerosis in lupus. JCI Insight 3, 99276 (2018).
Cole, S. et al. Integrative analysis reveals CD38 as a therapeutic target for plasma cell-rich pre-disease and established rheumatoid arthritis and systemic lupus erythematosus. Arthritis Res. Ther. 20, 85 (2018).
Orange, D. E. et al. Identification of three rheumatoid arthritis disease subtypes by machine learning integration of synovial histologic features and RNA sequencing data. Arthritis Rheumatol. 70, 690–701 (2018).
Walsh, A. M. et al. Triple DMARD treatment in early rheumatoid arthritis modulates synovial T cell activation and plasmablast/plasma cell differentiation pathways. PLOS ONE 12, e0183928 (2017).
Cuppen, B. V. J. et al. RNA sequencing to predict response to TNF-alpha inhibitors reveals possible mechanism for nonresponse in smokers. Expert Rev. Clin. Immunol. 14, 623–633 (2018).
Teitsma, X. M. et al. Identification of differential co-expressed gene networks in early rheumatoid arthritis achieving sustained drug-free remission after treatment with a tocilizumab-based or methotrexate-based strategy. Arthritis Res. Ther. 19, 170 (2017).
Ter Haar, N. M. et al. Reversal of sepsis-like features of neutrophils by interleukin-1 blockade in patients with systemic-onset juvenile idiopathic arthritis. Arthritis Rheumatol. 70, 943–956 (2018).
Mandelin, A. M. 2nd et al. Transcriptional profiling of synovial macrophages using minimally invasive ultrasound-guided synovial biopsies in rheumatoid arthritis. Arthritis Rheumatol. 70, 841–854 (2018).
Giladi, A. & Amit, I. Single-cell genomics: a stepping stone for future immunology discoveries. Cell 172, 14–21 (2018).
Landhuis, E. Single-cell approaches to immune profiling. Nature 557, 595–597 (2018).
Cheung, P., Khatri, P., Utz, P. J. & Kuo, A. J. Single-cell technologies — studying rheumatic diseases one cell at a time. Nat. Rev. Rheumatol. https://doi.org/10.1038/s41584-019-0220-z (2019).
Mizoguchi, F. et al. Functionally distinct disease-associated fibroblast subsets in rheumatoid arthritis. Nat. Commun. 9, 789 (2018).
Stephenson, W. et al. Single-cell RNA-seq of rheumatoid arthritis synovial tissue using low-cost microfluidic instrumentation. Nat. Commun. 9, 791 (2018).
Rao, D. A. et al. Pathologically expanded peripheral T helper cell subset drives B cells in rheumatoid arthritis. Nature 542, 110–114 (2017).
Kim, T. H., Choi, S. J., Lee, Y. H., Song, G. G. & Ji, J. D. Gene expression profile predicting the response to anti-TNF treatment in patients with rheumatoid arthritis; analysis of GEO datasets. Joint Bone Spine 81, 325–330 (2014).
Gaujoux, R. et al. Cell-centred meta-analysis reveals baseline predictors of anti-TNFα non-response in biopsy and blood of patients with IBD. Gut 68, 604–614 (2018).
Sweeney, T. E. et al. Unsupervised analysis of transcriptomics in bacterial sepsis across multiple datasets reveals three robust clusters. Crit. Care Med. 46, 915–925 (2018).
Zhang, F. et al. Defining inflammatory cell states in rheumatoid arthritis joint tissues by integrating single-cell transcriptomics and mass cytometry. Nat. Immunol. (in the press).
Arazi, A., R. D. et al. The immune cell landscape in kidneys of lupus nephritis patients. Nat. Immunol. (in the press).
Musters, A. et al. In rheumatoid arthritis, synovitis at different inflammatory sites is dominated by shared but patient-specific T cell clones. J. Immunol. 201, 417–422 (2018).
Sakurai, K. et al. HLA-DRB1 shared epitope alleles and disease activity are correlated with reduced T cell receptor repertoire diversity in CD4+T cells in rheumatoid arthritis. J. Rheumatol. 45, 905–914 (2018).
Kinslow, J. D. et al. Elevated IgA plasmablast levels in subjects at risk of developing rheumatoid arthritis. Arthritis Rheumatol. 68, 2372–2383 (2016).
Sakakibara, S. et al. Clonal evolution and antigen recognition of anti-nuclear antibodies in acute systemic lupus erythematosus. Sci. Rep. 7, 16428 (2017).
Lu, D. R. et al. T cell-dependent affinity maturation and innate immune pathways differentially drive autoreactive B cell responses in rheumatoid arthritis. Arthritis Rheumatol. 70, 1732–1744 (2018).
Elliott, S. E. et al. Affinity maturation drives epitope spreading and generation of proinflammatory anti-citrullinated protein antibodies in rheumatoid arthritis. Arthritis Rheumatol. 70, 1946–1958 (2018).
Titcombe, P. J. et al. Pathogenic citrulline-multispecific B cell receptor clades in rheumatoid arthritis. Arthritis Rheumatol. 70, 1933–1945 (2018).
Wang, J. J. et al. Molecular profiling and clonal tracking of secreted rheumatoid factors in primary Sjogren’s syndrome. Arthritis Rheumatol. 70, 1617–1625 (2018).
Gee, M. H. et al. Antigen identification for orphan T cell receptors expressed on tumor-infiltrating lymphocytes. Cell 172, 549–563 (2018).
Allis, C. D. & Jenuwein, T. The molecular hallmarks of epigenetic control. Nat. Rev. Genet. 17, 487–500 (2016).
Rivera, C. M. & Ren, B. Mapping human epigenomes. Cell 155, 39–55 (2013).
Alvarez-Errico, D., Vento-Tormo, R., Sieweke, M. & Ballestar, E. Epigenetic control of myeloid cell differentiation, identity and function. Nat. Rev. Immunol. 15, 7–17 (2015).
Smale, S. T., Tarakhovsky, A. & Natoli, G. Chromatin contributions to the regulation of innate immunity. Annu. Rev. Immunol. 32, 489–511 (2014).
Ivashkiv, L. B. & Park, S. H. Epigenetic regulation of myeloid cells. Microbiol. Spectr. https://doi.org/10.1128/microbiolspec.MCHD-0010-2015 (2016).
Wang, K. C. & Chang, H. Y. Epigenomics: technologies and applications. Circ. Res. 122, 1191–1199 (2018).
Ballestar, E. & Li, T. New insights into the epigenetics of inflammatory rheumatic diseases. Nat. Rev. Rheumatol. 13, 593–605 (2017).
Shi, L. et al. Monocyte enhancers are highly altered in systemic lupus erythematosus. Epigenomics 7, 921–935 (2015).
Zhang, Z. et al. H3K4 tri-methylation breadth at transcription start sites impacts the transcriptome of systemic lupus erythematosus. Clin. Epigenet. 8, 14 (2016).
Scharer, C. D. et al. ATAC-seq on biobanked specimens defines a unique chromatin accessibility structure in naive SLE B cells. Sci. Rep. 6, 27030 (2016).
Ai, R. et al. Comprehensive epigenetic landscape of rheumatoid arthritis fibroblast-like synoviocytes. Nat. Commun. 9, 1921 (2018).
Netea, M. G., Latz, E., Mills, K. H. & O’Neill, L. A. Innate immune memory: a paradigm shift in understanding host defense. Nat. Immunol. 16, 675–679 (2015).
Biswas, S. K. & Lopez-Collazo, E. Endotoxin tolerance: new mechanisms, molecules and clinical significance. Trends Immunol. 30, 475–487 (2009).
Saeed, S. et al. Epigenetic programming of monocyte-to-macrophage differentiation and trained innate immunity. Science 345, 1251086 (2014).
Park, S. H. et al. Type I interferons and the cytokine TNF cooperatively reprogram the macrophage epigenome to promote inflammatory activation. Nat. Immunol. 18, 1104–1116 (2017).
Novakovic, B. et al. Beta-glucan reverses the epigenetic state of LPS-induced immunological tolerance. Cell 167, 1354–1368 (2016).
Vatanen, T. et al. Variation in microbiome LPS immunogenicity contributes to autoimmunity in humans. Cell 165, 842–853 (2016).
Shi, L. et al. Endotoxin tolerance in monocytes can be mitigated by alpha2-interferon. J. Leukoc. Biol. 98, 651–659 (2015).
Wendeln, A. C. et al. Innate immune memory in the brain shapes neurological disease hallmarks. Nature 556, 332–338 (2018).
ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74 (2012).
Sanyal, A., Lajoie, B. R., Jain, G. & Dekker, J. The long-range interaction landscape of gene promoters. Nature 489, 109–113 (2012).
Ye, C. J. et al. Intersection of population variation and autoimmunity genetics in human T cell activation. Science 345, 1254665 (2014).
Raj, T. et al. Polarization of the effects of autoimmune and neurodegenerative risk alleles in leukocytes. Science 344, 519–523 (2014).
Lee, M. N. et al. Common genetic variants modulate pathogen-sensing responses in human dendritic cells. Science 343, 1246980 (2014).
Farh, K. K. et al. Genetic and epigenetic fine mapping of causal autoimmune disease variants. Nature 518, 337–343 (2015).
Raj, P. et al. Regulatory polymorphisms modulate the expression of HLA class II molecules and promote autoimmunity. eLife 5, e12089 (2016).
Martinez-Bueno, M. et al. Trans-ethnic mapping of BANK1 identifies two independent SLE-risk linkage groups enriched for co-transcriptional splicing marks. Int. J. Mol. Sci. 19, E2331 (2018).
Pulecio, J., Verma, N., Mejia-Ramirez, E., Huangfu, D. & Raya, A. CRISPR/Cas9-based engineering of the epigenome. Cell Stem Cell 21, 431–447 (2017).
Mumbach, M. R. et al. Enhancer connectome in primary human cells identifies target genes of disease-associated DNA elements. Nat. Genet. 49, 1602–1612 (2017).
Sokhi, U. K. et al. Dissection and function of autoimmunity-associated TNFAIP3 (A20) gene enhancers in humanized mouse models. Nat. Commun. 9, 658 (2018).
Liao, H. K. et al. In vivo target gene activation via CRISPR/Cas9-mediated trans-epigenetic modulation. Cell 171, 1495–1507 (2017).
Robertson, K. D. DNA methylation and human disease. Nat. Rev. Genet. 6, 597–610 (2005).
Doody, K. M., Bottini, N. & Firestein, G. S. Epigenetic alterations in rheumatoid arthritis fibroblast-like synoviocytes. Epigenomics 9, 479–492 (2017).
Hammaker, D. & Firestein, G. S. Epigenetics of inflammatory arthritis. Curr. Opin. Rheumatol. 30, 188–196 (2018).
Jin, B., Li, Y. & Robertson, K. D. DNA methylation: superior or subordinate in the epigenetic hierarchy? Genes Cancer 2, 607–617 (2011).
Mardis, E. R. Next-generation DNA sequencing methods. Annu. Rev. Genomics Hum. Genet. 9, 387–402 (2008).
Shendure, J. & Ji, H. Next-generation DNA sequencing. Nat. Biotechnol. 26, 1135–1145 (2008).
Feil, R., Charlton, J., Bird, A. P., Walter, J. & Reik, W. Methylation analysis on individual chromosomes: improved protocol for bisulphite genomic sequencing. Nucleic Acids Res. 22, 695–696 (1994).
Lister, R. et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature 462, 315–322 (2009).
Reinders, J. & Paszkowski, J. Bisulfite methylation profiling of large genomes. Epigenomics 2, 209–220 (2010).
Meissner, A. et al. Reduced representation bisulfite sequencing for comparative high-resolution DNA methylation analysis. Nucleic Acids Res. 33, 5868–5877 (2005).
Kurdyukov, S. & Bullock, M. DNA methylation analysis: choosing the right method. Biology 5, E3 (2016).
Nakano, K., Whitaker, J. W., Boyle, D. L., Wang, W. & Firestein, G. S. DNA methylome signature in rheumatoid arthritis. Ann. Rheum. Dis. 72, 110–117 (2013).
Whitaker, J. W. et al. An imprinted rheumatoid arthritis methylome signature reflects pathogenic phenotype. Genome Med. 5, 40 (2013).
Ai, R. et al. DNA methylome signature in synoviocytes from patients with early rheumatoid arthritis compared to synoviocytes from patients with longstanding rheumatoid arthritis. Arthritis Rheumatol. 67, 1978–1980 (2015).
Frank-Bertoncelj, M. et al. Epigenetically-driven anatomical diversity of synovial fibroblasts guides joint-specific fibroblast functions. Nat. Commun. 8, 14852 (2017).
Rhead, B. et al. Rheumatoid arthritis naive T cells share hypermethylation sites with synoviocytes. Arthritis Rheumatol. 69, 550–559 (2017).
Mok, A. et al. Hypomethylation of CYP2E1 and DUSP22 promoters associated with disease activity and erosive disease among rheumatoid arthritis patients. Arthritis Rheumatol. 70, 528–536 (2018).
Chung, S. A. et al. Genome-wide assessment of differential DNA methylation associated with autoantibody production in systemic lupus erythematosus. PLOS ONE 10, e0129813 (2015).
Mok, A. et al. Genome-wide profiling identifies associations between lupus nephritis and differential methylation of genes regulating tissue hypoxia and type 1 interferon responses. Lupus Sci. Med. 3, e000183 (2016).
Cole, M. B. et al. Epigenetic signatures of salivary gland inflammation in Sjogren’s syndrome. Arthritis Rheumatol. 68, 2936–2944 (2016).
Puliti, A., Caridi, G., Ravazzolo, R. & Ghiggeri, G. M. Teaching molecular genetics: chapter 4—positional cloning of genetic disorders. Pediatr. Nephrol. 22, 2023–2029 (2007).
The International FMF Consortium. Ancient missense mutations in a new member of the RoRet gene family are likely to cause familial Mediterranean fever. Cell 90, 797–807 (1997).
Zhu, X. et al. Whole-exome sequencing in undiagnosed genetic diseases: interpreting 119 trios. Genet. Med. 17, 774 (2015).
Zhou, Q. et al. Loss-of-function mutations in TNFAIP3 leading to A20 haploinsufficiency cause an early onset autoinflammatory syndrome. Nat. Genet. 48, 67–73 (2016).
Zappala, Z. & Montgomery, S. B. Non-coding loss-of-function variation in human genomes. Hum. Hered. 81, 78–87 (2016).
Ma, M. et al. Disease-associated variants in different categories of disease located in distinct regulatory elements. BMC Genomics 16, S3 (2015).
Tesi, B. et al. A RAB27A 5ʹ untranslated region structural variant associated with late-onset hemophagocytic lymphohistiocytosis and normal pigmentation. J. Allergy Clin. Immunol. 142, 317–321 (2018).
Namjou, B. et al. Evaluation of the TREX1 gene in a large multi-ancestral lupus cohort. Genes Immun. 12, 270–279 (2011).
Beaudoin, M. et al. Deep resequencing of GWAS loci identifies rare variants in CARD9, IL23R and RNF186 that are associated with ulcerative colitis. PLOS Genet. 9, e1003723 (2013).
Cardinale, C. J. et al. Targeted resequencing identifies defective variants of decoy receptor 3 in pediatric-onset inflammatory bowel disease. Genes Immun. 14, 447 (2013).
Nakagawa, K. et al. Somatic NLRP3 mosaicism in Muckle-Wells syndrome. A genetic mechanism shared by different phenotypes of cryopyrin-associated periodic syndromes. Ann. Rheumat. Dis. 74, 603–610 (2015).
Tanaka, N. et al. High incidence of NLRP3 somatic mosaicism in patients with chronic infantile neurologic, cutaneous, articular syndrome: results of an international multicenter collaborative study. Arthritis Rheum. 63, 3625–3632 (2011).
Zhou, Q. et al. Cryopyrin-associated periodic syndrome caused by a myeloid-restricted somatic NLRP3 mutation. Arthritis Rheumatol. 67, 2482–2486 (2015).
Holzelova, E. et al. Autoimmune lymphoproliferative syndrome with somatic Fas mutations. N. Engl. J. Med. 351, 1409–1418 (2004).
Savola, P. et al. Somatic mutations in clonally expanded cytotoxic T lymphocytes in patients with newly diagnosed rheumatoid arthritis. Nat. Commun. 8, 15869 (2017).
Rowczenio, D. M. et al. Late-onset cryopyrin-associated periodic syndromes caused by somatic NLRP3 mosaicism—UK single center experience. Front. Immunol. 8, 1410 (2017).
Yuri, K. et al. Identification of a high-frequency somatic NLRC4 mutation as a cause of autoinflammation by pluripotent cell–based phenotype dissection. Arthritis Rheumatol. 69, 447–459 (2017).
Chung, J. et al. The minimal amount of starting DNA for Agilent’s hybrid capture-based targeted massively parallel sequencing. Sci. Rep. 6, 26732 (2016).
Grossman, R. L. et al. Toward a shared vision for cancer genomic data. N. Engl. J. Med. 375, 1109–1112 (2016).
Acknowledgements
The work of L.T.D., L.B.I. and K.-H.P.-M. was supported by grants from the US National Institutes of Health (NIH).
Reviewer information
Nature Reviews Rheumatology thanks P. Gaffney and the other anonymous reviewers for their contribution to the peer review of this work.
Author information
Authors and Affiliations
Contributions
All authors researched data for the article, provided substantial contributions to discussions of content and wrote the article. L.T.D., A.I. and L.B.I. reviewed and/or edited the manuscript before submission.
Corresponding author
Ethics declarations
Competing interests
R.M.S. declares that he is an employee of Novartis. The other authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
AMP RA and SLE network: https://amp-ralupus.stanford.edu/about/ra-lupus-amp-project/
Gene Expression Omnibus: https://www.ncbi.nlm.nih.gov/geo/
Human Cell Atlas: https://www.humancellatlas.org/
ImmPort: https://www.immport.org/home/
International Human Epigenome Consortium: http://ihec-epigenomes.org/welcome/
PRECISESADS: http://www.precisesads.eu/
The LifeTime Initiative: https://lifetime-fetflagship.eu/
Rights and permissions
About this article
Cite this article
Donlin, L.T., Park, SH., Giannopoulou, E. et al. Insights into rheumatic diseases from next-generation sequencing. Nat Rev Rheumatol 15, 327–339 (2019). https://doi.org/10.1038/s41584-019-0217-7
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41584-019-0217-7
This article is cited by
-
Evaluation and correlation analysis of ocular surface disorders and quality of life in autoimmune rheumatic diseases: a cross-sectional study
BMC Ophthalmology (2023)
-
Inching closer to precision treatment for rheumatoid arthritis
Nature Medicine (2022)
-
Synovial Tissue: Cellular and Molecular Phenotyping
Current Rheumatology Reports (2019)