Article Text
Abstract
Objectives Treatment of rheumatic diseases requires immunomodulatory agents which can compromise antibody production. However, even in case of agents directly targeting B cells, a minority of patients develop hypogammaglobulinaemia, suggesting a genetic predisposition, which has not been investigated so far. The phenotypic overlap between primary immunodeficiency disorders (PIDs) and rheumatic diseases suggests a shared genetic basis, especially in case of patients with rheumatic diseases with hypogammaglobulinaemia.
Methods 1008 patients with rheumatic diseases visiting the outpatient clinics of the Hannover University Hospital were screened for hypogammaglobulinaemia. Those with persistent hypogammaglobulinaemia and an equal number of patients without it underwent targeted next-generation sequencing, searching for variations in genes linked with hypogammaglobulinaemia in the context of PIDs.
Results We identified 33 predicted pathogenic variants in 30/64 (46.9%) patients with persistent secondary hypogammaglobulinaemia. All 33 variants were monoallelic and 10 of them in 10/64 (15.6%) patients were found in genes associated with autosomal dominant PIDs. 2/64 (3.1%) patients harboured variants which were previously reported to cause PIDs. In the group without hypogammaglobulinaemia we identified seven monoallelic variants in 7/64 (10.9%), including a variant in a gene associated with an autosomal dominant PID.
Conclusions Approximately half of patients with persistent secondary hypogammaglobulinaemia harboured at least a variant in a PID gene. Despite the fact that previous immunomodulatory treatment is an exclusion criterion in the diagnosis of PIDs, we identified genetic variants that can account for PID in patients with clear rheumatic phenotypes who developed hypogammaglobulinaemia after the introduction of immunomodulatory treatment. Our data suggest the common genetic causes of primary and secondary hypogammaglobulinaemia.
- secondary hypogammaglobulinemia
- primary immunodeficiency
- common variable immunodeficiency
- inborn errors of immunity
- dmard
Statistics from Altmetric.com
- secondary hypogammaglobulinemia
- primary immunodeficiency
- common variable immunodeficiency
- inborn errors of immunity
- dmard
Key messages
What is already known about this subject?
Immunomodulatory agents for the treatment of rheumatic diseases induce hypogammaglobulinaemia in a minority of treated, suggesting a likely genetic predisposition.
What does this study add?
This is the first study evaluating the genetic background of secondary hypogammaglobulinaemia.
Despite the fact that primary immunodeficiency is most often conceived as susceptibility to infections, identification of variants in primary immunodeficiency disorder (PID) genes in a cohort of patients with rheumatic diseases, most of whom had no history of severe or recurrent infections, suggests that rheumatic disease may be the dominant phenotypic aspect of PID.
How might this impact on clinical practice or future developments?
This study questions the classification of hypogammaglobulinaemia into primary and secondary, especially in patients with rheumatic diseases, alerting treating physicians for considering PID in patients with rheumatic diseases with hypogammaglobulinaemia.
The latter may lead to re-evaluation of treatment of hypogammaglobulinaemia in patients with rheumatic diseases and consideration of precision-directed therapies, which are employed to treat autoimmune manifestations of monogenic PIDs.
Introduction
To diagnose primary hypogammaglobulinaemia, and especially common variable immunodeficiency (CVID), secondary causes of hypogammaglobulinaemia need to be excluded.1 These include protein-losing conditions, haematological malignancies and certain medications, such as anticonvulsive and immunomodulatory agents.2 Some of the latter target B cells directly, whereas others have a broader impact on the immune system, which nonetheless affects B cells and antibody production. Treatment with immunomodulatory agents accounts for secondary immunodeficiency and hypogammaglobulinaemia in patients with rheumatic diseases. In addition to recurrent infections, the phenotype of CVID includes autoimmunity, also in the form of a rheumatic disease, such as rheumatoid arthritis (RA), systemic lupus erythematosus and Sjögren’s syndrome (SjS).3–5 As autoimmune manifestations in the context of CVID usually necessitate treatment with immunosuppressive agents, the discrimination between primary and secondary hypogammaglobulinaemia can become challenging, especially when immunoglobulin (Ig) levels before treatment introduction have not been controlled.6 7
Expanding evidence, and especially the discovery and phenotypic characterisation of monogenic primary immunodeficiency disorder (PID), suggests that primary immunodeficiency and autoimmunity share common pathogenic pathways.8 The advent of next-generation sequencing (NGS) aided the identification of PID-causing genetic defects. The spectrum of genetic variants underlying PID is expanding and currently, defects in more than 400 genes have been linked to PID.9 The most common symptomatic adult-onset PID, CVID,10 is largely a polygenic disease, though considering the more recent reports, the proportion of monogenic forms appears to expand, exceeding 20% of cases.11 12 However, incomplete penetrance and variable expressivity of genetic variants reported as PID-causing, question the exact division between monogenic and polygenic forms and suggest the influence of additional genetic modifiers, epigenetic regulation and/or environmental factors.13 The phenotypes of relatively common monogenic PIDs, such as due to NFKB1 loss-of-function variants, STAT3 gain-of-function (GOF) variants, CTLA4 insufficiency, LRBA deficiency and activated PI3K delta syndrome include features of rheumatic diseases such as arthritis, enthesiopathy and vasculitis.14–20
As discussed above, immunomodulatory regimens for rheumatic diseases can lead to hypogammaglobulinaemia.2 However, even after introduction of rituximab, which directly targets B cells and reduces plasma cell precursors, only a minority of treated patients develop hypogammaglobulinaemia,21 22 suggesting a genetic vulnerability or even a genetic cause of secondary hypogammaglobulinaemia. Despite recent advances in understanding the genetic basis of primary immunodeficiency, evidence on the genetics of secondary immunodeficiency is scarce. The phenotypic overlap between adult-onset primary antibody deficiencies and secondary hypogammaglobulinaemia in the context of rheumatic diseases suggests shared genetic aetiologies. Hence, we employed a panel NGS-approach searching for primary hypogammaglobinaemia-associated variants in a cohort of patients with rheumatic diseases and persistent hypogammaglobulinaemia, developing after introduction of treatment with immunomodulatory agents.
Patients and methods
Study cohort
This single-centre study included all patients with rheumatic disease visiting our rheumatology outpatient clinic between November 2018 and March 2019 (N=1008, figure 1). Visits of patients were scheduled approximately every 3–6 months and serum Ig levels were measured at every visit. The normal range of serum IgG values for adults lies between 7 g/L and 16 g/L. Secondary hypogammaglobulinaemia has been defined as persistently reduced IgG (<7 g/L) at time of the study and in follow-up visits during at least the year before the study, developing after the introduction of immunomodulatory regimens including prednisolone, diverse synthetic and/or biological disease-modifying antirheumatic drugs (DMARD), known to cause hypogammaglobulinaemia,2 in patients who previously had normal or high IgG levels.
Study design.
Targeted NGS
Blood samples were collected in the outpatient clinics of the department of Rheumatology and Immunology of Hannover University Hospital. Genomic DNA (gDNA) was isolated from peripheral whole blood using QIAamp DNA Blood Midi Kit, according to the manufacturer’s protocol (Qiagen). Targeted NGS was performed with a gene panel (Agilent Technologies), comprising known and candidate genes associated with primary antibody deficiencies (online supplemental table 1), using a MiSeq desktop sequencer (Illumina) as described previously.23 The detected genetic alterations were validated by Sanger sequencing using a service from Eurofins. We analysed the original NGS data with Agilent SureCall software (Agilent Technologies). Genome Reference Consortium Human Build 37 was employed as reference genome. Allele frequency, variant annotation and potential functional effect were considered for variant selection. Variants with an allele frequency in the general population higher that 1% according to the Genome Aggregation Database were not considered. The functional effect of nonsense, frameshift, splice site affecting or start/stop codon introducing variants was evaluated with the following bioinformatics tools: Combined Annotation-Dependent Depletion (CADD) Score, Mutation Taster, Protein Variation Effect Analyser and Polymorphism Phenotyping v2.
Supplemental material
Statistical analysis
For statistical calculation we used GraphPad prism 8 (GraphPad, La Jolla, USA). Descriptive statistics are reported as median and IQR in case of continuous variables and as counts and percentages for dichotomous variables. Categorical variables were compared by the Χ2 test. Non-categorical variables were compared with the Mann-Whitney U test. To correct for multiple testing, p values were adjusted for Benjamini-Hochberg false discovery rate (FDR). P values were considered significant if they were lower than a threshold selected to control an FDR of 5%.
Results
Characteristics of patients with rheumatic diseases with hypogammaglobulinaemia
Out of 72 identified patients with rheumatic diseases with persistent hypogammaglobulinaemia, secondary to treatment with immunomodulatory agents, 64 were enrolled in the study (figure 1). The rest (ie, 8/72 patients) were excluded, due to lack of consent or gDNA. The 64 patients with secondary hypogammaglobulinaemia and an equal number of randomly selected patients with rheumatic diseases with normal serum IgG levels underwent targeted NGS. The characteristics of all 128, who underwent NGS as well as those of patients from the original cohort, are summarised in table 1. Most patients (52/64, 81.3%) had an isolated reduction of IgG. In addition to reduced IgG, 6/64 (9.4%) patients had low IgA, 6/64 (9.4%) had low IgM and one patient displayed panhypogammaglobulinaemia. Most patients were diagnosed with hypogammaglobulinaemia while receiving conventional synthetic DMARDs (online supplemental table 2). Retrospective evaluation of medical records of studied patients with hypogammaglobulinaemia revealed recurrent or severe infections in 15/64 (23.4%) of them. These were mostly recurrent upper respiratory tract infections. One patient had recurrent skin abscesses and 2/64 (3.1%) had a history of recurrent herpes reactivations necessitating antiviral prophylaxis. Further, 2/64 (3.1%) patients had a history of candida esophagitis but none of them had recurrent candida or other fungal infections and none was receiving prophylactic antifungals. Six of sixty-four (9.4%) patients were receiving prophylactic antibiotics and 8/64 (12.5%) of them were on Ig replacement therapy.
Patients’ characteristics and immunomodulatory regimes
Variants in autosomal dominant PID-causing genes
The employed targeted NGS approach included a panel of genes linked to predominantly antibody deficiencies (online supplemental table 1). Considering allele frequency as well as the CADD and Mutation Significance Cut-off scores of each identified variant, as described above, we ended up with 35 rare and likely deleterious variations in 31/64 patients (48.4%), all of which were monoallelic (figure 2).
Summary of genetic findings in n=64 patients with rheumatic diseases with secondary hypogammaglobulinaemia; each green cycle matches a studied patient. Boxes indicate genetic findings for each patient classified as monoallelic variants in genes associated with autosomal dominant (AD) primary immunodeficiency disorders (PID) (marked with blue colour) or autosomal recessive (AR) primary PID (marked with yellow colour). Monoallelic variants in TNFRSF13B are indicated with orange boxes and those in CLEC16A with green ones. Diagnosis of rheumatic disease is indicated in the left side of the graph. RA, rheumatoid arthritis; SjS, Sjögren’s syndrome; SLE, systemic lupus erythematosus; SpA, spondyloarthritis.
Ten patients had a variant in a gene linked to autosomal dominant (AD) PID (figure 3). In particular, five patients harboured variants in NFKB1, the gene encoding the p105 subunit of the transcription factor NF-κB1. Three of four identified NFKB1 variants (table 2) were missense variants and the c.865G>T variant, which was identified in two unrelated patients, is predicted to be a stop codon-gain variant. Among the identified NFKB1 variants, the c.1601G>A (p.R534H) substitution has been previously reported to cause monogenic CVID.13 Further, we identified a rare missense variant in PIK3CD in patient with RA, a PTEN missense variant in a patient with spondyloarthritis (SpA), one nonsense variant in NFKBIA in a patient with RA and a missense variant in STAT3 in a patient with late-onset RA. The latter variant has been previously reported to be a GOF, resulting in PID.15 Clinical and immunological characteristics of patients harbouring variants in AD PID-causing genes are summarised in online supplemental table 3 and online supplemental table 4, respectively.
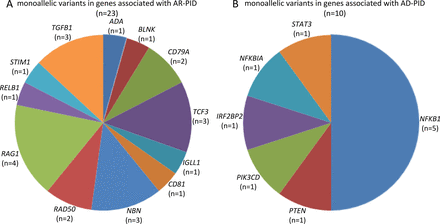
Summary of genes, whose variations were detected in a cohort of c patients with rheumatic diseases with hypogammaglobulinaemia developing after introduction of immunomodulatory regimens, (A) genes linked to AR-PID and (B) genes linked to AD-PID. AD-PID, autosomal dominant primary immunodeficiency disorder; AR-PID, autosomal recessive PID.
Monoallelic variants in genes associated with autosomal dominant PIDs, identified in patients with rheumatic diseases with persistent hypogammaglobulinaemia and those without
Half of patients with secondary hypogammaglobulinaemia had a predominantly articular rheumatic disease (32/64), diagnosed as either RA (18/64) or SpA (14/64). Articular disease was more common among patients with variants in AD PID-causing genes (8/10 vs 2/54, p=0.0389, q=0.0648). The prevalence of infectious manifestations was similar in patients with variants in AD PID-causing genes and those without (3/10 vs 13/54, p=0.691, q=0.838). With respect to the immunological parameters of studied patients with hypogammaglobulinaemia, concomitant reduction of IgA and/or IgM appeared with same frequency among patients with hypogammaglobulinaemia with or without variants in AD-PID genes (2/10 vs 11/54, p=1, q=1). Further, retrospective evaluation of available peripheral lymphocyte subset counts revealed that patients with a variant in AD-PID genes displayed significantly lower proportions of class-switched (online supplemental text).
Monoallelic variants in TNFRSF13B, CLEC16A and autosomal recessive PID-causing genes
In addition, we detected 23 variants in genes associated with autosomal recessive (AR) PID in 21/64 patients with secondary hypogammaglobulinaemia, which only in the context of homozygosity or compound heterozygosity would cause a PID. These included variants in genes linked to AR-PIDs, including agammaglobulinaemia (CD79A, TCF3, BLNK and IGLL1) and severe combined immunodeficiency genes (ADA, RAG1, STIM1) (figure 3, table 3). Variants in TNFRSF13B, the gene encoding the transmembrane activator and calcium-modulating cyclophilin ligand interactor, have been reported in a considerable proportion of patients with CVID and are rather predisposing but do not solely cause hypogammaglobulinaemia.24 25 Among the 64 tested patients, 3 (4.7%) had a rare monoallelic variant in TNFRSF13B (table 3). Considering the previously reported association of CLEC16A single-nucleotide polymorphism with CVID and the reported B cell dysfunction in Clec16 knockdown mice,26 which both suggest the pathogenic relevance of CLEC16A in PID, we tested our patients for CLEC16A variants and identified three different monoallelic missense variants in 3/64 patients with secondary hypogammaglobulinaemia (table 3).
Monoallelic variants in genes associated with autosomal recessive PIDs as well as in CLEC16A and TNFRSF13B, identified in patients with rheumatic diseases with persistent hypogammaglobulinaemia and those without
Targeted NGS in patients with rheumatic diseases without hypogammaglobulinaemia
To evaluate the association of the above described genetic findings with secondary hypogammaglobulinaemia in the context of rheumatic disease rather than with the rheumatic disease itself, in parallel to the 64 patients with hypogammaglobulinaemia we tested an equal number of patients with rheumatic diseases without hypogammaglobulinaemia. Seeking for variants in the same PID-related genes, we identified 6 AR-PID variants (table 3) and a single AD-PID variant (table 2) in 7/64 (10.9%) patients with rheumatic diseases without hypogammaglobulinaemia. Both AR-PID and AD-PID variants were more commonly detected among patients with rheumatic diseases with hypogammaglobulinaemia than those without (patients with at least one AR-PID variant: 21/64 vs 6/64, p=0.0012, q=0.007; patients with at least one AD-PID variant: 10/64 vs 1/64, p=0.0045, q=0.015). The AD-PID variant was found in IRF2BP2 (c.958C>A, p.P320T), in a woman with SjS and recurrent herpes infections (table 2, online supplemental file 1 and online supplemental table 4). Considering the fact that immunodeficiency due to heterozygous IRF2BP2 mutations does not necessarily cause hypogammaglobulinaemia,27 the identified variant may account for recurrent herpes infections in this patient.
Retrospective evaluation of medical records of the 128 sequenced patients revealed that 47/64 with persistent hypogammaglobulinaemia and 27/64 without hypogammaglobulinaemia had received no corticosteroids or other immunomodulatory agent at first presentation in our outpatient clinic. Evaluation of IgG values at first presentation of those treatment-naive patients revealed similar IgG levels between patients harbouring at least one genetic variant in a PID-related gene and those without any PID variant (see online supplemental text and online supplemental figure 1).
Discussion
While the spectrum of genetic defects underlying CVID is expanding,8 the genetic basis of secondary hypogammaglobulinaemia remained unknown. Here, in a cohort of patients with diverse rheumatic diseases, who developed hypogammaglobulinaemia after introduction of an immunomodulatory therapy, we identified at least a variant in a PID-associated gene in approximately half (48.4%) of studied patients. This finding suggests an at least partially shared genetic background for primary and secondary hypogammaglobulinaemia. Further, we show that a sizeable minority of patients with predominantly articular rheumatic diseases, that is, RA and SpA, harboured genetic variants, which could account for hypogammaglobulinaemia in the context of PID, even leading to reclassification of physician-diagnosed rheumatic disease into PID. It is noteworthy that the identified variants, especially in AD-PID genes, are predicted to be deleterious but were not functionally tested to evaluate their pathogenicity. However, two variants in AD-PID genes were previously reported to cause CVID-like immunodeficiency. Identification of variants in AR PID-related genes does not explain hypogammaglobulinaemia in patients with rheumatic diseases. Nonetheless, the fact that such variants were more often detected among patients with rheumatic diseases with hypogammaglobulinaemia, suggests their representing risk factors for hypogammaglobulinaemia, which needs to be further investigated in larger cohorts of patients with rheumatic disease.
PID may manifest as autoimmune disease, necessitating treatment with immunomodulatory agents that can induce hypogammaglobulinaemia, independently of the underlying PID. Immunological investigations and especially the measurement of Ig levels as well as the documentation of infections before and after starting an immunomodulatory treatment can aid differentiating a pre-existing hypogammaglobulinaemia or susceptibility to infections, falling under PID, from a secondary immunodeficiency. However, considering the natural history of primary hypogammaglobulinaemia and its likely progressive course,28 clinically evident immunodeficiency may follow the onset of autoimmunity and therefore, the introduction of a hypogammaglobulinaemia-inducing immunomodulatory treatment. In that case, the differentiation of primary from secondary hypogammaglobulinaemia can be challenging or even impossible. The identification of genetic variants that could account for PID in a cohort of patients with rheumatic diseases and per se secondary hypogammaglobulinaemia, developing after introduction of immunomodulatory regimens, suggests that genetic testing, may be of diagnostic value in resolving the above-presented diagnostic dilemma between primary and secondary hypogammaglobulinaemia.
Timely distinguishing of PID from secondary hypogammaglobulinaemia may be relevant in clinical practice. Diagnosis of an underlying PID results in a higher degree of vigilance for the identification of infections, malignancies, such as gastric cancer and lymphoproliferative diseases,29 30 which may be relevant for patients with rheumatic diseases harbouring variants in PID-associated genes. Identification of the overlapping genetic background of primary and secondary hypogammaglobulinaemia in rheumatic disease may lead to the expansion of the indication for Ig replacement, which is currently limited to PID,31 also for patients with rheumatic diseases with recurrent secondary hypogammaglobulinaemia-associated infections. Further, detection of genetic variants conferring risk for hypogammaglobulinaemia in patients with rheumatic diseases may result in extra caution before introducing immunomodulatory regimens with a relatively stronger immunosuppressive or hypogammaglobulinaemia-inducing effect, such as glucocorticoids or rituximab.21 32 33 Finally, patients with rheumatic diseases harbouring particular defects such as STAT3 or PIC3CD GOF variants or CTLA-4 insufficiency may benefit from individualised therapeutic approaches, already employed to treat autoimmune manifestations in the context of PID.34–37 However, parameters such as the penetrance of disease-causing variants and the natural history of each monogenic condition, as well as the availability and the cost of genetic testing, need to be determined before launching routine screening of patients with rheumatic diseases for PID variants prior to starting immunomodulatory therapies.
Despite the fact that immunodeficiency is most often conceived as susceptibility to infectious diseases, identification of PID-causing mutations in patients with rheumatic diseases, highlights the fact that PID may manifest with autoimmunity. The term inborn errors of immunity (IEI), is a synonym for ‘primary immune deficiency disorders’, which highlights the increasingly identified genetic background of PIDs.8 38 These conditions are monogenic defects, whose phenotypic description is largely based on cohorts of patients with clinically evident immunodeficiency, that is, infectious manifestations. Our identification of PID-causing variants in patients with rheumatic diseases without noticeable infection records, suggests that autoimmunity and immune dysregulation can be the dominant phenotypic aspect of IEI. Especially the identification of monoallelic variants in AD PID-associated genes in a cohort of patients with rheumatic diseases, suggests that phenotypic characterisation of IEI based on cohorts of patients with clinically evident immunodeficiency may overestimate the prevalence of infectious manifestations at the expense of autoimmune phenotypes.
Our study has several limitations. As discussed above, despite using stringent selection criteria, including the rarity and pathogenicity prediction scores, we did not demonstrate the pathogenicity of identified variants, which may lead to an overestimation of the incidence of PID-related hypogammaglobulinaemia among patients with rheumatic diseases. In addition, despite testing for variations in genes most commonly accounting for IEI, such as NFKB1, STAT3, CTLA4 and PIK3CD, the employed panel did not include all genes previously associated with primary hypogammaglobulinaemia, which would be possible through whole exome sequencing. Considering the expanding number of gene defects reported to be involved in PID, it is likely that a subset of tested patients with rheumatic diseases may harbour hypogammaglobulinaemia-causing variants in genes which were not included in our gene panel.
In summary, the identification genetic variants that can account for PID in patients with clear rheumatic phenotypes who developed hypogammaglobulinaemia after the introduction of immunomodulatory agents provides evidence on the overlapping genetic aetiology of primary and secondary hypogammaglobulinaemia in the context of rheumatic diseases. Our data suggest that primary immunodeficiency and autoimmune rheumatic diseases are not mutually exclusive entities, but rather related pathophysiological processes.
Acknowledgments
We thank all nurses, physicians and documentation personnel of the outpatient clinics of the department of Rheumatology and Immunology of the Hannover Medical School for collecting blood samples, informing the patients about the study and documenting patients’ medications.
References
Supplementary materials
Supplementary Data
This web only file has been produced by the BMJ Publishing Group from an electronic file supplied by the author(s) and has not been edited for content.
Footnotes
Handling editor Josef S Smolen
Contributors GS and FA conceived and planned the study. GS took the lead in writing the manuscript. TW and RES significantly contributed to drafting and revision of the paper. GS and FA contributed substantially to data acquisition and interpretation, and performed the statistical analysis. ND, IRA and MA collected DNA samples and performed targeted NGS. All authors approved the final version.
Funding This project was funded by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) under Germany’s Excellence Strategy—EXC 2155 'RESIST'—Project ID 39087428 and the German network for multi-organ autoimmune diseases (GAIN). GS receives funding from the Young Academy Clinician/Scientist program of Hannover Medical School, Germany and the Rosemarie-Germscheid foundation. IRA receives funding from the German Academic Exchange Service (DAAD), the Hannover Biomedical Research School (HBRS) and the Center for Infection Biology (ZIB). All authors and this project are supported by the German Center for Infection Research (DZIF TTU 07.801).
Competing interests None declared.
Patient and public involvement Patients and/or the public were not involved in the design, or conduct, or reporting, or dissemination plans of this research.
Patient consent for publication All studied patients signed an informed consent form.
Ethics approval This study was conducted in accordance with the Declaration of Helsinki and was also approved from the Ethical Committee of the Hannover Medical School (approval number: 8875).
Provenance and peer review Not commissioned; externally peer reviewed.
Data availability statement Data are available upon reasonable request. Data are available for formal research purposes only upon request to the corresponding author.